Material:
Ni3S
ID:
MMD-1652
Navigation:
Space group | Lattice parameters | Thermodynamic properties | Phase diagram | Magnetic properties | Atomic positions and site-specific magnetic data | Pair-wise magnetic data | Methods | References
Space group:
Crystal system |
hexagonal |
Space group number |
194 |
Hermann-Mauguin |
P6_3/mmc |
Hall |
-P 6c 2c |
Point group |
6/mmm |
Structure data:
Normalized formula |
Ni3S |
The number of formula units per unit cell |
2 |
The total number of atoms per unit cell |
8 |
The number of inequivalent sites per unit cell |
2 |
Structure search |
MP |
Lattice parameters:
a (Å) |
5.0859 |
b (Å) |
5.0859 |
c (Å) |
4.0159 |
α (deg.) |
90.000 |
β (deg.) |
90.000 |
γ (deg.) |
120.000 |
Volume (Å3) |
89.961 |
Density (g/cm3) |
7.684 |
Crystal structure visualization:
Thermodynamic properties:
| DFT calculations (details) | |
|---|---|
Formation energy (vs. elemental phases) |
152.1 meV/atom |
Formation energy above hull |
378.2 meV/atom |
Phase diagram:
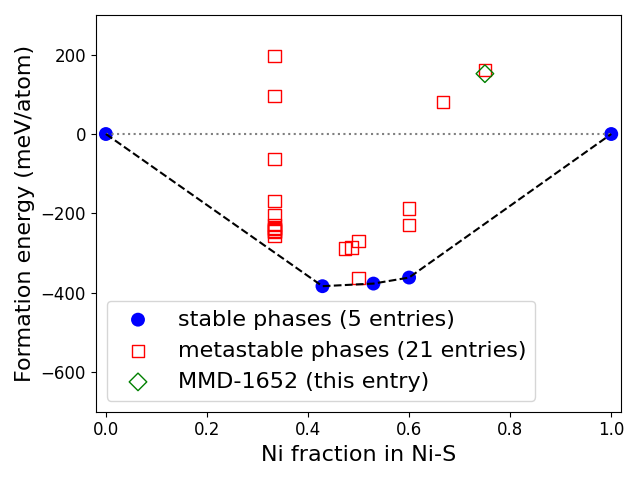
Related structures:
Compounds with the same formula: Ni3S |
2 entries found |
Compounds with the same elements: Ni-S |
25 entries found |
Magnetic properties:
| DFT calculations (details) | |
|---|---|
Magnetic ordering |
Ferromagnetic |
Total magnetic moment |
2.85 μB/cell |
Averaged magnetic moment |
0.36 μB/atom |
Magnetic polarization, Js = μ0Ms |
0.37 T (= 294.4 emu/cm3) |
| LMTO-GF calculations (details) | |
|---|---|
Curie temperature, TC |
|
| DFT calculations (details) | |
|---|---|
Magnetic anisotropy constant, Ka-c |
0.58 MJ/m3 (= 0.33 meV/cell) |
Magnetic easy axis |
c |
Magnetic hardness parameter, κ |
2.38 |
Atomic positions (fractional coordinates) and site-specific magnetic data:
| index | species | w | x | y | z | m (μB) | Esoc100 (meV) | Esoc001 (meV) |
|---|---|---|---|---|---|---|---|---|
| 1 | Ni | 6h | 0.166858 | 0.333716 | 0.250000 | 0.47 | . | . |
| 2 | Ni | 6h | 0.666284 | 0.833142 | 0.250000 | 0.47 | . | . |
| 3 | Ni | 6h | 0.166858 | 0.833142 | 0.250000 | 0.47 | . | . |
| 4 | Ni | 6h | 0.833142 | 0.666284 | 0.750000 | 0.47 | . | . |
| 5 | Ni | 6h | 0.333716 | 0.166858 | 0.750000 | 0.47 | . | . |
| 6 | Ni | 6h | 0.833142 | 0.166858 | 0.750000 | 0.47 | . | . |
| 7 | S | 2d | 0.333333 | 0.666667 | 0.750000 | 0.07 | . | . |
| 8 | S | 2d | 0.666667 | 0.333333 | 0.250000 | 0.07 | . | . |
m: local magnetic moment
Esoc100 (001): Spin-orbit coupling energy for the magnetization oriented along the crystallographic a (c) axis
(Individual Esoc values are temporarily not available while we perform maintenance.)
Site-resolved magnetic moments:
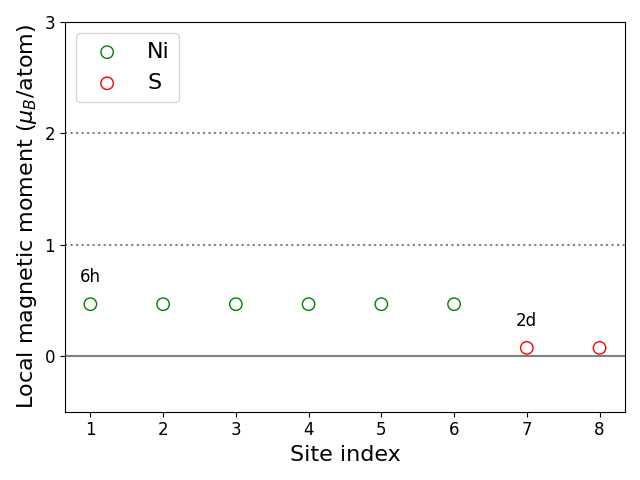
Pair-wise magnetic data:
| site i | site j | distance (Å) | Jij (meV) | ||||
|---|---|---|---|---|---|---|---|
| 1 | Ni | 6h | 2 | Ni | 6h | 2.54 | . |
| 1 | Ni | 6h | 3 | Ni | 6h | 2.54 | . |
| 1 | Ni | 6h | 4 | Ni | 6h | 3.56 | . |
| 1 | Ni | 6h | 5 | Ni | 6h | 2.49 | . |
| 1 | Ni | 6h | 6 | Ni | 6h | 2.49 | . |
| 1 | Ni | 6h | 7 | S | 2d | 2.49 | . |
| 1 | Ni | 6h | 8 | S | 2d | 2.54 | . |
| 2 | Ni | 6h | 3 | Ni | 6h | 2.54 | . |
| 2 | Ni | 6h | 4 | Ni | 6h | 2.49 | . |
| 2 | Ni | 6h | 5 | Ni | 6h | 3.56 | . |
| 2 | Ni | 6h | 6 | Ni | 6h | 2.49 | . |
| 2 | Ni | 6h | 7 | S | 2d | 2.49 | . |
| 2 | Ni | 6h | 8 | S | 2d | 2.54 | . |
| 3 | Ni | 6h | 4 | Ni | 6h | 2.49 | . |
| 3 | Ni | 6h | 5 | Ni | 6h | 2.49 | . |
| 3 | Ni | 6h | 6 | Ni | 6h | 3.56 | . |
| 3 | Ni | 6h | 7 | S | 2d | 2.49 | . |
| 3 | Ni | 6h | 8 | S | 2d | 2.54 | . |
| 4 | Ni | 6h | 5 | Ni | 6h | 2.54 | . |
| 4 | Ni | 6h | 6 | Ni | 6h | 2.54 | . |
| 4 | Ni | 6h | 7 | S | 2d | 2.54 | . |
| 4 | Ni | 6h | 8 | S | 2d | 2.49 | . |
| 5 | Ni | 6h | 6 | Ni | 6h | 2.54 | . |
| 5 | Ni | 6h | 7 | S | 2d | 2.54 | . |
| 5 | Ni | 6h | 8 | S | 2d | 2.49 | . |
| 6 | Ni | 6h | 7 | S | 2d | 2.54 | . |
| 6 | Ni | 6h | 8 | S | 2d | 2.49 | . |
| 7 | S | 2d | 8 | S | 2d | 3.56 | . |
Export data (under construction)
Terms and conditions
- csv
- json
- yaml
Pair-resolved magnetic exchange parameters:
Diagram is not available for this entry.
Individual exchange parameters:
Diagram is not available for this entry.
Methods:
DFT calculations |
|
LMTO-GF calculations |
|
References:
References |
Materials Project: mp-976881 |
You can download and use the data of this database for your scientific work, provided that you express proper acknowledgements:
-
M. Sakurai, R. Wang, T. Liao, C. Zhang, H. Sun, Y. Sun, H. Wang, X. Zhao, S. Wang, B. Balasubramanian, X. Xu, D. J. Sellmyer, V. Antropov, J. Zhang, C.-Z. Wang, K.-M. Ho, J. R. Chelikowsky,
"Discovering rare-earth-free magnetic materials through the development of a database," [selected as Editors' Suggestion]
Phys. Rev. Materials 4, 114408 (2020).
DOI: 10.1103/PhysRevMaterials.4.114408 [BibTex, RIS] -
B. Balasubramanian, M. Sakurai, C.-Z. Wang, X. Xu, K.-M. Ho, J. R. Chelikowsky, and D. J. Sellmyer,
"Synergistic computational and experimental discovery of novel magnetic materials," [Review article]
Mol. Syst. Des. Eng., 5, 1098-1117 (2020).
DOI: 10.1039/D0ME00050G [BibTex, RIS] - Dataset as of March 2020.
-
Portal page: https://www.novomag.physics.iastate.edu/structure-database
- See references for original papers about specific alloys listed on this site.